Note: This post was updated to work with NetworkX and for clarity.
It’s possible this will turn out like the day when Python 2.5 introduced [coroutines][coroutines]. At the time I was very excited. I spent several hours trying to convince my coworkers we should immediately abandon all our existing Java infrastructure and port it to finite state machines implemented using Python coroutines. After a day of hand waving over a proof of concept, we put that idea aside and went about our lives.
Soon after, I left for a Python shop, but in the next half decade I still never found a good place to use this interesting feature.
But it doesn’t feel like that.
As I come to terms more with switching to Python 3.2, the futures module seems similarly exciting. I wish I’d had it years ago, and it’s almost reason in itself to upgrade from Python 2.7. Who cares if none of your libraries have been ported yet?
This library lets you take any function and distribute it over a process pool. To test that out, we’ll generate a bunch of random graphs and iterate over all their cliques.
Code
First, let’s generate some test data using the dense_gnm_random_graph function. Our data includes 1000 random graphs, each with 100 nodes and 100 * 100 edges.
import networkx as nx
n = 100
graphs = [nx.dense_gnm_random_graph(n, n*n) for _ in range(1000)]
Now we write a function iterate over all cliques in a given graph. NetworkX provides a find_cliques function which returns a generator. Iterating over them ensures we will run through the entire process of finding all cliques for a graph.
def iterate_cliques(g):
for _ in nx.find_cliques(g):
pass
Now we just define two functions, one for running in serial and one for running in parallel using futures.
from concurrent import futures
def serial_test(graphs):
for g in graphs:
iterate_cliques(g)
def parallel_test(graphs, max_workers):
with futures.ProcessPoolExecutor(max_workers=max_workers) as executor:
executor.map(iterate_cliques, graphs)
Our __main__ simply generates the random graphs, samples from them, times both functions, and write CSV data to standard output.
from csv import writer
import random
import sys
import time
if __name__ == '__main__':
out = writer(sys.stdout)
out.writerow(['num graphs', 'serial time', 'parallel time'])
n = 100
graphs = [nx.dense_gnm_random_graph(n, n*n) for _ in range(1000)]
# Run with a number of different randomly generated graphs
for num_graphs in range(50, 1001, 50):
sample = random.choices(graphs, k = num_graphs)
start = time.time()
serial_test(sample)
serial_time = time.time() - start
start = time.time()
parallel_test(sample, 16)
parallel_time = time.time() - start
out.writerow([num_graphs, serial_time, parallel_time])
The output of this script shows that we get a fairly linear speedup to this code with little effort.
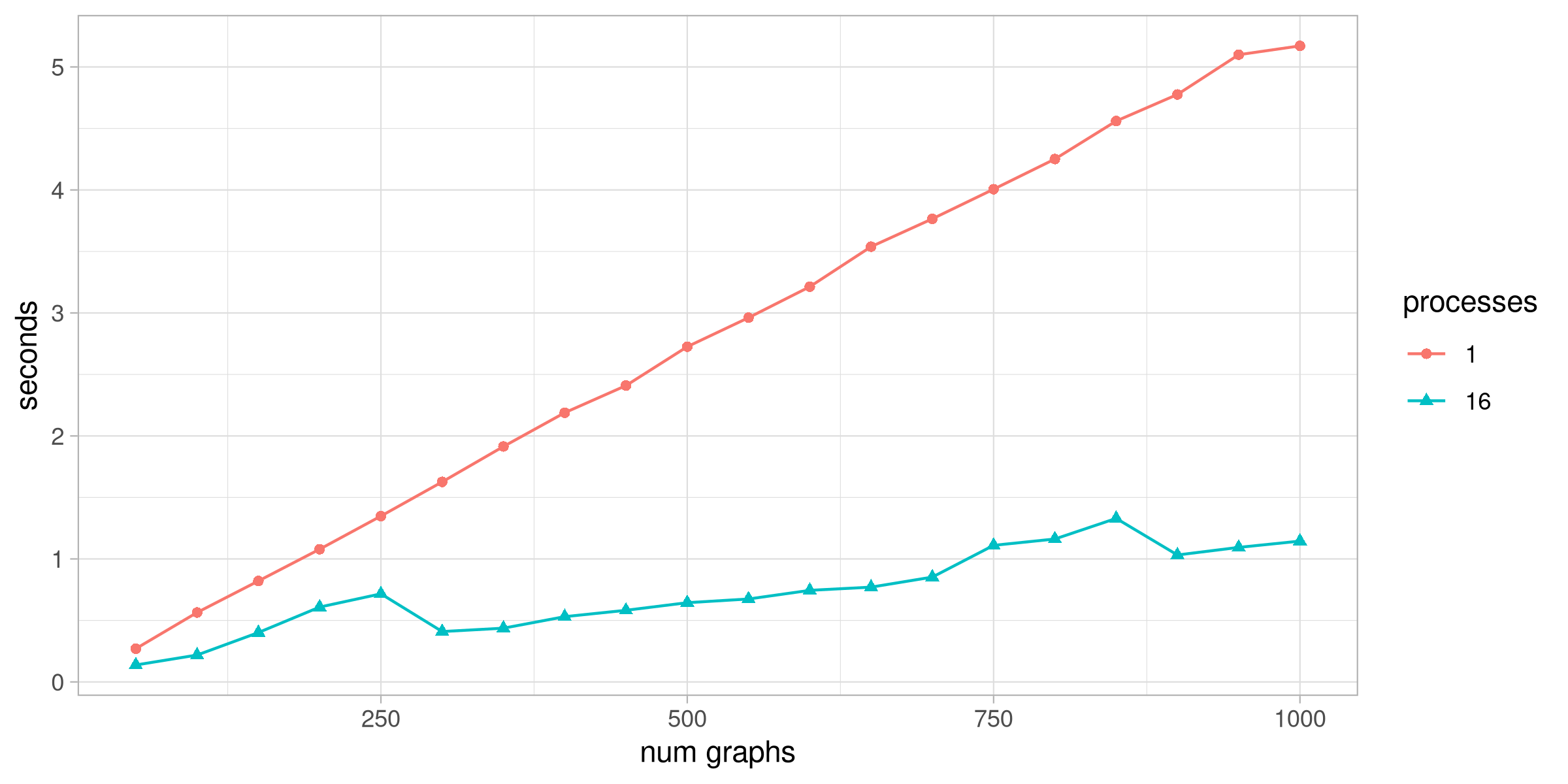
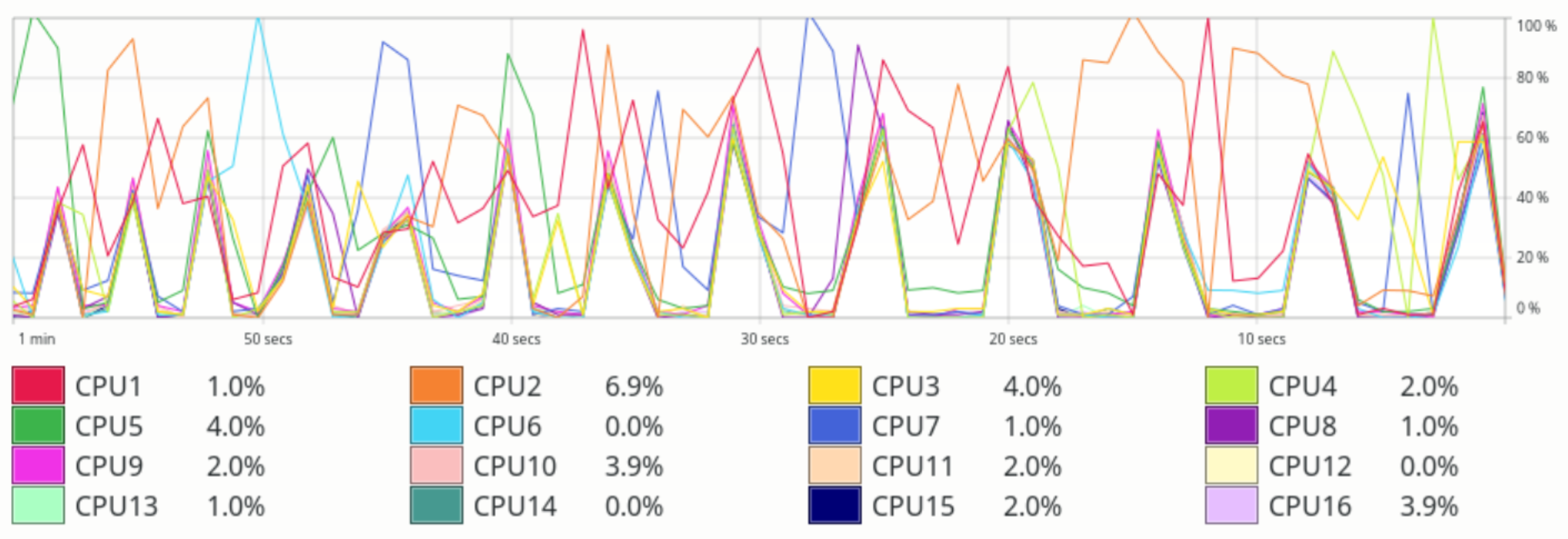
I ran this on a machine with 8 cores and hyperthreading. Eyeballing the chart, it looks like the speedup is roughly 5x. My system monitor shows spikes on CPU usage across cores whenever the parallel test runs.
Resources
- Output data
- Full source listing